SeqPig is a library for Apache Pig http://pig.apache.org/ for the distributed analysis of large sequencing datasets on Hadoop clusters. With SeqPig one can easily take advantage of the advanced high-level features of Pig to manipulate and analyze sequencing data, thus writing simple scripts that automatically run as scalable distributed programs on potentially very large Hadoop clusters.
SeqPig provides a number of features and functionalities. It provides import and export functions for file formats commonly used in bioinformatics, as well as a collection of Pig user-defined functions (UDF's) specifically designed for processing aligned and unaligned sequencing data. Currently SeqPig supports BAM, SAM, FastQ and Qseq input and output and FASTA input, thanks in part to the functionality provided by the Hadoop-BAM library.
This document can also be found under http://seqpig.sourceforge.net/.
Contact information: mailto:seqpig-users@lists.sourceforge.net
Releases of SeqPig come bundled with Picard/Samtools, which is developed at the Wellcome Trust Sanger Institute, and Seal, which is developed at CRS4. For more details on these projects see their respective web sites, at http://picard.sourceforge.net/, http://samtools.sourceforge.net/ and http://biodoop-seal.sourceforge.net/.
Since SeqPig builds on top of Pig, which itself relies on Hadoop for job execution, the installation requires a working Pig and Hadoop setup. For more information in this direction see for example http://pig.apache.org/docs/r0.11.1/start.html. A convenient way to install Hadoop is provided by the the Cloudera Hadoop distribution (CDH).
We have tested SeqPig with the following dependencies.
The following two dependencies are already included in a pre-compiled SeqPig release:
On a Cloudera Hadoop installation with Pig a suitable environment configuration would be:
This way, you'll be able to start a SeqPig-enabled Pig shell by running the seqpig command.
Note that since the sources are included in the release you are also able to build SeqPig if necessary, as described below.
For more information on how to build Seal see http://biodoop-seal.sourceforge.net/installation.html.
Note: the Picard and Sam jar files are contained in the Hadoop-BAM release for convenience.
Once you've built SeqPig, you can move the directory to a location of your preference (if on a shared system, perhaps /usr/local/java/seqpig, else even your home directory could be fine).
Some of the example scripts in this manual (e.g., Section 3.2.6) require functions from PiggyBank, which is a collection of publicly available User-Defined Functions (UDF's) that are distributed with Pig but may need to be built separately, depending on your Pig distribution. For more details see https://cwiki.apache.org/confluence/display/PIG/PiggyBank. Verify that PiggyBank has been compiled by looking for the file piggybank.jar under $PIG_HOME:
If PiggyBank hasn't been compiled, go into $PIG_HOME/contrib/piggybank/java and run ant.
Assuming you have started an interactive Pig Job Flow (for example via the AWS console), you can login into the master node and copy the SeqPig release to the Hadoop user home directory. Then set both SEQPIG_HOME and PIG_HOME correctly (HADOOP_HOME should be set by default). Note that the Pig version installed does not necessarily match the latest Pig release. The advantage, however, is the ability to use S3 buckets for input and output.
Consider the following example for starting SeqPig on Amazon Elastic MapReduce. In this example we install SeqPig into /home/hadoop/seqpig.
After building SeqPig it may be a good idea to run tests to verify that the environment has been set up correctly. When inside the SeqPig directory execute
By default the tests run Pig in local mode. In order to test Hadoop mode pass the command line argument -h. Note that this test requires that Hadoop to be set up correctly. It first imports a BAM file, sorts the reads by coordinate and converts it to SAM for comparing the results. The test should end with the line
If you intend to run SeqPig on an Amazon Elastic MapReduce instance, you can also test input from S3 by providing an S3 path to the file data/input.bam:
for example:
where seqpig is the name of the S3 bucket.
Assuming that all the environment variables have been set as described in the previous sections, you can start the SeqPig-enabled “grunt” shell by running
If you prefer to tun SeqPig in local mode (without Hadoop), which can be useful for debugging scripts, you can start it by running
Alternatively to using the interactive Pig grunt shell, users can write scripts that are then submitted to Pig/Hadoop for automated execution. This type of execution has the advantage of being able to handle parameters; for instance, one can parametrize input and output files. See the /scripts directory inside the SeqPig distribution and Section 3 for examples.
This section lists a number of examples of the types of operations that can be performed with SeqPig. Each example is typically a mix of standard Pig operations and SeqPig user-defined functions (UDF's). Note that once the data has been imported to Pig the only difference between, say, read data originating from BAM and Fastq files, is that some fields in tuples derived from BAM may not be available in those from Fastq, e.g., alignment start positions.
To access sequencing data in BAM files, SeqPig uses Hadoop-BAM, which provides access to all fields and optional attributes in the data records. All the examples below assume that an input BAM file is initially imported to HDFS via
This additional setup step is required to extract the header and store it inside HDFS to simplify some operations that only operate on a chunk of the BAM file without having access to the original header. Similarly, for uncompressed SAM files there is a corresponding prepareSamInput.sh. Once the input file is imported into HDFS it can be loaded in the grunt shell via
The 'yes' parameter to BamLoader chooses read attributes to be loaded; choose 'no' whenever these are not required, which is also the default value).
Once some operations have been performed, the resulting (possibly modified) read data can then be stored into a new BAM file via
and can also be exported from HDFS to the local filesystem via
Writing the BAM data to the screen (similarly to samtools view) can be done simply by
Another very useful Pig command is describe, which returns the schema that Pig uses for a given data bag. Example:
returns
Notice that all fields except the attributes are standard data types (strings or integers). Specific attributes can be accessed via attributes#'name'. For example,
will output all read names and their corresponding MD tag. Other useful commands are LIMIT and SAMPLE, which can be used to select a subset of reads from a BAM/SAM file.
will assign the first 20 records of A to B, while
will sample from A with sampling probability 0.01.
Since the flags field of a SAM record is exposed to Pig, one can simply use it to filter out all tuples (i.e., SAM records) that do not have the corresponding bit set.
For convenience SeqPig provides a set of filters that allow a direct access to the relevant fields. The previous example is equivalent to
Note that the above example assumes that the Pig shell was started in the SeqPig root directory. If this is not the case, you need to adjust the path to filter_defs.pig accordingly. For of a full list of the available filters look at this file.
Other fields can also be used for filtering, for example the read mapping quality value as shown below.
SeqPig also supports filtering by samtools region syntax. The following examples selects base positions 1 to 44350673 of chromosome 20.
Note that filtering by regions requires a valid SAM header for mapping sequence names to sequence indices. This file is generated automatically when BAM files are imported via the prepareBamInput.sh script.
Sorting an input BAM file by chromosome, reference start coordinate, strand and readname (in this hierarchical order):
Note that strand here is used to refer to the strand flag computed by the expression (flags/16)%2.
Computing read coverage over reference-coordinate bins of a fixed size, for example:
will output the number of reads that lie in any non-overlapping bin of size 200 base pairs. For a more precise coverage computation see Section 3.1.8 on computing read pileup.
Generating samtools compatible pileup (for a correctly sorted BAM file with MD tags aligned to the same reference, should produce the same output as samtools mpileup -A -f ref.fasta -B input.bam):
The script essentially does the following:
There are two optional parameters for pileup.pig: min_map_qual and min_base_qual (both with default value 0) that filter out reads with either insufficient map quality or base qualities. Their values can be set the same way as the other parameters above.
There is an alternative pileup script which typically performs better but is more sensitive to additional parameters. This second script, pileup2.pig, is based on a binning of the reads according to intervals on the reference sequence. The pileup output is then generated on a by-bin level and not on a by-position level. This script can be invoked with the same parameters as pileup2.pig. However, it has tunable parameters that determine the size of the bins (binsize) and the maximum number of reads considered per bin (reads_cutoff), which is similar to the maximum depth parameter that samtools accepts. However, note that since this parameter is set on a per-bin level you may choose it dependent on the read length and bin size, as well as the amount of memory available on the compute nodes.
In order to evaluate the output of an aligner, it may be useful to consider the distribution of the mapping quality over the collection of reads. Thanks to Pig's GROUP operator this is fairly easy.
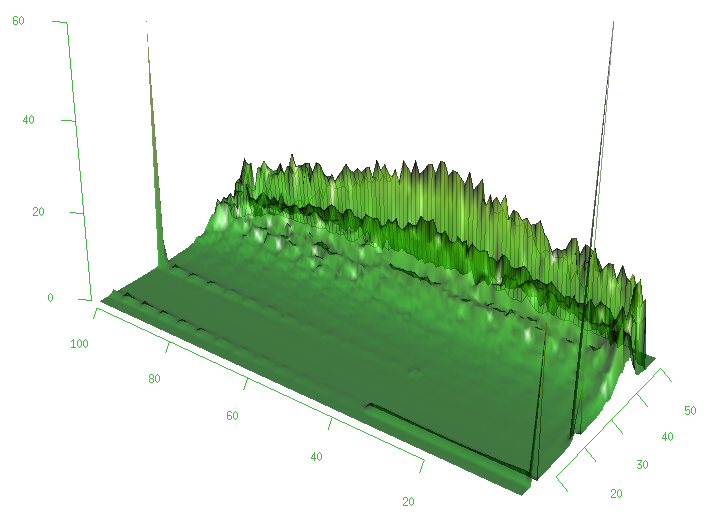
Sometimes it may be useful to analyze a given set of reads for a bias towards certain bases being called at certain positions inside the read. The following simple script generates for each reference base and each position inside a read the distribution of the number of read bases that were called.
Here is an example output (for a BAM file with 50 reads):
Figure 1 shows the distribution obtained from a sample BAM file.

Analogously to the previous example collecting statistics for the read bases, we can also collect frequencies for base qualities conditioned on the position of the base inside the reads. If these fall off too quickly for later positions, it may indicate some quality issues with the run. The resulting script is actually fairly similar to the previous one with the difference of not grouping over the reference bases.
Here is an example output (for a BAM file with 50 reads):
The script filter_mappability.pig filters reads in a given BAM file based on a given mappability threshold. Both input BAM and mappability file need to reside inside HDFS.
Note that since the script relies on distributing the BAM file header and the mappability file via Hadoop's distributed cache, it is not possible to run it with Pig in local mode.
SeqPig supports the import and export of non-aligned reads stored in Qseq and Fastq data. Due to Pig's model that all records correspond to tuples, which form bags, reads can be processed in very much the same way independent of, for example, whether they are stored in Qseq or Fastq. Also, since Fastq and Qseq files are header-less, there is no pre-processing step required prior to loading the files, compared to SAM/BAM import.
The following two lines simply convert an input Qseq file into Fastq format.
The other direction works analogously.
The following examples are not specific to the type of input data (Fastq, Qseq, BAM, …). They roughly correspond to the ones computed by the popular FastQC tool and can be all found in the fast_fastqc.pig script.
Once a read set is imported, it is fairly straightforward to compute simple statistics, such as a histogram of the length of reads. Using the LENGTH string function, which is part of the PiggyBank (see Section 2.4.1), we can first compute the set of read lengths, group them by their value and then compute the size of each of the groups.
By using a SeqPig UDF to split reads into bases, one can compute this statistic by relying on Pig's GROUP BY operator as follows.
Alternatively, SeqPig also provides another UDF that has better performance for larger read sets.
The following example shows how one can exploit Pig's GROUP and COUNT operators to obtain statistics of the composition of read bases.
Similarly to GC content, one can rely on Pig operations to compute for each read position the distribution of nucleotides and their base qualities, as given by the aligner. However, in order to improve performance for larger data sets, SeqPig also provides UDF's for this purpose.
Assuming there were some problems in certain cycles of the sequencer, it may be useful to clip bases from reads. This example removes the last 3 bases and their qualities and stores the data under a new filename. Note that here we rely on the SUBSTRING and LENGTH string functions, which are part of the PiggyBank (see Section 2.4.1).
Recent releases of Pig (such as 0.11.0) have a parameter that enables aggregation of partial results on the mapper side, which greatly improves performances of some of the UDF's described above. Further, another option that is worth considering is to use so-called schema tuples. For more information see the Pig documentation. The recommended setting is as follows:
Some scripts (such as pileup2.pig) require an amount of memory that depends on the choice of command line parameters. To tune the performance of such operations on the Hadoop cluster, consider the following Hadoop-specific parameters in mapred-site.xml.
Additionally, it is possible to pass command line parameters (such as mapper and reducer memory limits). For instance, consider the Pig invocation (see tools/tun_all_pileup2.sh)
By setting appropriate values for MAP_MEMORY, REDUCE_MEMORY, CHILD_MEMORY and for the number of available reduce slots REDUCE_SLOTS one may be able to improve performance.
For optimal performance and space usage it may be advisable to enable the compression of Hadoop map (and possibly reduce) output, as well as temporary data generated by Pig. Compression with Pig can be enabled by setting properties such as
on the Pig command line or the Hadoop configuration. Note that currently not all Hadoop compression codecs are supported by Pig. The details regarding which compression codec to use depend are beyond the scope of this manual.
The aim of this section is to give a brief overview of the available UDF's that come with SeqPig, their input and output schemas and their operation. For more detailed information please consult the source code.
The ReadRefPositions UDF takes an aligned read and outputs a bag of tuples, one for each base, that gives the position on the reference sequence this base corresponds to.
| Usage: | |||
Field | Type | Description |
|
Input schema: | read | chararray | read bases |
flags | integer | SAM flags |
|
chr | chararray | reference name / chromosome |
|
position | integer | mapping position |
|
cigar | chararray | CIGAR string |
|
basequal | chararray | base qualities |
|
Output schema:
(bag) | chr | charray | reference name / chromosome |
pos | integer | position (on reference) |
|
base | chararray | read base |
|
qual | chararray | base quality |
|
This package contains import and export functions for the various file formats that are supported. In the case of BAM and SAM files, the file formats make use of all the fields that are available for a Picard sequence record (SAMRecord).
The BAM and SAM loader output schemas are identical and both loaders take an optional argument that can take values “yes” and “no”, indicating whether the map of attributes should be populated. Reading in attributes incurs an additional overhead during the import of the data, so choose “no” whenever these are not needed. For a proper definition of the various fields see the SAM file format description available at http://samtools.sourceforge.net/SAMv1.pdf.
| Usage: | |||
Output schema: | Field | Type | Description |
name | chararray | read name |
|
start | integer | start position of alignment |
|
end | integer | end position of alignment |
|
read | chararray | read bases |
|
cigar | charray | CIGAR string |
|
basequal | chararray | base qualities |
|
flags | integer | flags |
|
insertsize | integer | insert size |
|
mapqual | integer | mapping quality |
|
matestart | integer | mate start position |
|
materefindex | integer | mate reference index |
|
refindex | integer | reference index |
|
refname | chararray | reference name |
|
attributes | map | SAMRecord attributes |
|
The BAM and SAM storer input schemas are identical and both are essentially equal to the output schema of the corresponding loader functions. Note that the order of the fields inside tuples does not matter, only their field names need to be present. Both loaders require, however, the name of a file that contains a valid SAM header. For details see Section 3.1.
| Usage: | |||
Input schema: | Field | Type | Description |
name | chararray | read name |
|
start | integer | start position of alignment |
|
end | integer | end position of alignment |
|
read | chararray | read bases |
|
cigar | charray | CIGAR string |
|
basequal | chararray | base qualities |
|
flags | integer | flags |
|
insertsize | integer | insert size |
|
mapqual | integer | mapping quality |
|
matestart | integer | mate start position |
|
materefindex | integer | mate reference index |
|
refindex | integer | reference index |
|
refname | chararray | reference name |
|
attributes | map | SAMRecord attributes |
|
Both loaders for unaligned read file formats Fastq and Qseq essentially provide the same output schema for the tuple field names they produce. Note that some fields that are not present in the input data may remain empty.
| Usage: | |||
Output schema: | Field | Type | Description |
instrument | chararry | instrument identifier |
|
run_number | integer | run number |
|
flow_cell_id | chararray | flow cell ID |
|
lane | integer | lane number |
|
tile | integer | tile |
|
xpos | integer | XPos |
|
ypos | integer | YPos |
|
read | integer | read ID |
|
qc_passed | boolean | QC flag |
|
control_number | integer | control number |
|
index_sequence | chararray | index sequence |
|
sequence | chararray | read bases |
|
quality | chararray | base qualities |
|
The Fastq and Qseq storer input schemas are identical and both are essentially equal to the output schema of the corresponding loader functions. Note that the order of the fields inside tuples does not matter, only their field names need to be present. All fields except sequence and quality are optional.
| Usage: | |||
Input schema: | Field | Type | Description |
instrument | chararry | instrument identifier |
|
run_number | integer | run number |
|
flow_cell_id | chararray | flow cell ID |
|
lane | integer | lane number |
|
tile | integer | tile |
|
xpos | integer | XPos |
|
ypos | integer | YPos |
|
read | integer | read ID |
|
qc_passed | boolean | QC flag |
|
control_number | integer | control number |
|
index_sequence | chararray | index sequence |
|
sequence | chararray | read bases |
|
quality | chararray | base qualities |
|
The FastaLoader loads reference sequence data in FASTA format and produces tuples that correspond to segments of the reference. These tuples could then be, for example, co-grouped with tuples that result from an imported BAM file of reads aligned to a matching reference sequence in order to compare read with reference bases. For more information on the FASTA format see http://en.wikipedia.org/wiki/FASTA_format.
| Usage: | |||
Output schema: | Field | Type | Description |
index_sequence | chararray | name of reference fragment (e.g., chromX) |
|
start | integer | 1-based start position of current reference fragment |
|
sequence | chararray | reference bases |
|
This module contains various functions that implement Apache Pig's filter UDF interface.
The CoordinateFilter filters aligned reads by samtools regions. See also Section 3.1.4. Note that this filter requires a SAM file header to operate, which can be extracted for example by running prepareBamInput.sh script when uploading a BAM file to HDFS.
| Usage: | |||
Input schema: | Field | Type | Description |
refindex | integer | reference sequence index |
|
start | integer | start position |
|
end | integer | end position |
|
SAMFlagsFilter is a filter that uses SAM flags for filtering. For more example of filtering by SAM flags see Section 3.1.2. Also note the pre-defined set of filters that can be activated by running run scripts/filter_defs.pig from within the SeqPig main directory.
| Usage: | |||
Input schema: | Field | Type | Description |
flags | integer | SAM flags of read |
|
The BaseFilter is not actually a filter UDF in the Pig sense. Instead, it scans the given set of 2-tuples, consisting of read bases and their base qualities, and replaces all bases that have quality values below a given threshold by “N”.
| Usage: | |||
Input schema: | Field | Type | Description |
sequence | chararray | read bases |
|
quality | integer | base qualities |
|
This package contains some UDF's that aim to provide FastQC like statistics for sequence data. They rely on an optimized counter class that maintains counters for discrete range variables (nucleotides, base qualities, etc.) that can be expressed as pairs. For example, one may be interested in counting the number of times base quality 30 appears at position 10 within a given set of reads. This type of counter implements interfaces of Apache Pig that, among other things, allow for in-memory map side aggregation of subresults, which leads to significant runtime improvements due to a reduction in memory allocation overhead. This optimization comes at the expense of fixing the set of possible values that can be observed (bases, base qualities) and the number of position in question (the readlength). In the rest of the section we therefore assume a given upper bound on the readlength, which we denote by READLENGTH. For implementation details of the counter see ItemCounter2D.java.
The BaseCounts UDF relies on the aforementioned counter class to provide a histogram of the occurences of each of the five base identifiers (“A”, “C”, “G”, “T” and “N”) over the given set of reads. Note that the output consists of an array of Java long values that are encoded inside a byte array. For convenience one should call the UDF FormatQualCounts to unpack the byte arrays and obtain the original counter values.
| Usage: | |||
Field | Type | Description |
|
| Input schema: | arbitrary | chararray | bases |
| Output schema: | long_array_NxM | bytearray | counter values encoded as bytes (N=5 and M is equal to READLENGTH) |
FormatBaseCounts takes the output of BaseCounts and extracts its counter values. For a complete example see above.
| Usage: | |||
Field | Type | Description |
|
| Input schema: | long_array_NxM | bytearray | counter values encoded as bytes (N=5 and M is equal to READLENGTH) |
Output schema:
(bag) | pos | long | position (ranging from 0 to READLENGTH-1) |
count1…qualN | double | fraction of reads having this base at the given position |
|
The AvgBaseQualCounts UDF relies on the aforementioned counter class to provide a histogram of the average base quality (rounded to next integer) over the given set of reads (average taken over the positions within the read). Note that the output consists of an array of Java long values that are encoded inside a byte array. For convenience one should call the UDF FormatAvgBaseQualCounts to unpack the byte arrays and obtain the original counter values.
| Usage: | |||
Field | Type | Description |
|
| Input schema: | arbitrary | chararray | base qualities |
| Output schema: | long_array_NxM | bytearray | counter values encoded as bytes (N is the number of different quality values and M is 1 in this case) |
FormatAvgBaseQualCounts takes the output fo AvgBaseQualCounts and extracts its counter values. For a complete example see above.
| Usage: | |||
Field | Type | Description |
|
| Input schema: | long_array_NxM | bytearray | counter values encoded as bytes (N is the number of different quality values and M is 1 in this case) |
Output schema:
(bag) | pos | long | position (always 1 in this case) |
mean | double | avg. base quality, averaged over the set of reads |
|
stdev | double | standard deviation of average base quality over the set of reads |
|
qual1…qualN | double | number of occurences of each (average) quality |
|
| Usage: | |||
Field | Type | Description |
|
| Input schema: | arbitrary | chararray | base qualities |
| Output schema: | long_array_NxM | bytearray | counter values encoded as bytes (N is the number of different quality values and M is the READLENGTH) |
FormatBaseQualCounts takes the output fo BaseQualCounts and extracts its counter values. For a complete example see above.
| Usage: | |||
Field | Type | Description |
|
| Input schema: | long_array_NxM | bytearray | counter values encoded as bytes (N is the number of different quality values and M is the READLENGTH) |
Output schema:
(bag) | pos | long | position (ranging from 0 to READLENGTH-1) |
mean | double | avg. base quality at this position, averaged over the set of reads |
|
stdev | double | standard deviation of base quality at this position |
|
qual1…qualN | double | number of occurences of each read quality at this position |
|
The pileup operation implemented in SeqPig basically consists of two stages. First, each read is inspected and pileup relevant output is produced for each of the positions of the reference that it “spans”. Note that this may be also more or less than the readlength, depending if there were insertions or deletions. Then the output of this first stage is grouped by (chromosome, position) and pileup output from different reads affecting the same reference position is merged. The output should be the same that can be obtained by an operation of samtools mpileup.
The last UDF in this section (BinReadPileup) increases parallelism by adding a pre-processing step that first partition reads into bins by their alignment position and then calls the two other UDF's in each of the bins. Fore more details see Section 3.1.8 and for a complete example see script scripts/pileup2.pig.
The ReadPileup UDF performs the steps of the first stage described above and operates on single reads that are passed with a subset of their fields. It produces a bag of tuples as output.
The PileupOutputFormatting UDF performs the second stage of the pileup, which is the merging of the output coming from different reads for the same reference position. Note that the UDF assumes that input is passed in the order of reference position. For usage see the example code above.
Field | Type | Description |
|
Input schema: | pileup | bag | bag of tuples produced by ReadPileup, grouped by position |
position | integer | reference position this pileup data originates from |
|
Output schema:
(bag) | refbase | charray | reference base |
count | integer | number of reads (depth) |
|
pileup | chararray | pileup string |
|
qual | chararray | base qualities |
This UDF aims to increase parallelism during the pileup computation by partitioning reads into bins by their alignment position and then calling the two previous UDF's in each of the bins. Note that each bin corresponds to a half-interval over the reference position of some reference segment (chromosome).